December 19, 2025
Designing Enzymes with AI? Why You Need a Physics-AI Hybrid for R&D Success
The biotech world witnessed a remarkable moment in 2024 when the Nobel Prize in Chemistry recognized two sides of a revolution in how we view protein structures. David Baker won half the prize for computational protein design, which applied physics-based methods to build entirely new proteins. The other half was shared by Demis Hassabis and John Jumper for developing AlphaFold2, an AI model that predicts complex protein structures with unprecedented accuracy.
For those working in enzyme development, these advances encapsulate the tension shaping the field. AI offers speed. Physics promises understanding. Your investment needs both, but interpretability still matters. As the buzz around AI and machine learning grows, the technology still struggles to predict chemical activity.
The hype is real, and so is the pressure to adopt before you’re left behind. But speed without interpretability is still just expensive guesswork. Below, we outline some of the realities behind choosing to pursue an AI-only method to enzyme development as well as some practical advantages of combining it with a physics-based approach.
The Trap of Black-Box AI
Let's start with what AI actually does in enzyme design. It's pattern recognition — sophisticated and powerful, but fundamentally limited by what’s been learned from past data. Think of it like predicting flight routes by analyzing historical records. AI sees patterns, identifies correlations, and suggests a way forward based on what worked before.
This approach has delivered genuine breakthroughs. AlphaFold2 revolutionized protein structure prediction because it processed massive amounts of high-quality training data from decades of work. When you have thousands of known protein structures, AI can learn the rules well enough to predict new ones with impressive accuracy.
But enzymes aren't just proteins. They're catalysts performing chemical reactions, and that changes everything. Each enzyme-substrate pair has unique chemistry, and the reaction mechanisms vary. To predict chemical activity with AI alone, you'd need enormous datasets for each type of reaction — data that simply doesn't exist yet. It's not a question of if AI will get there. It's only a question of when. However, right now, that "when" is still years away.
Here's the problem this creates: when a black-box AI model recommends an enzyme, you get a sequence. That's it. You have no explanation of why this sequence should work and no visibility into what makes this enzyme different from the thousands of other candidates.
If it works, great. But if it fails, you can't troubleshoot at the molecular level. You can't systematically improve the design. You're back to the trial-and-error screening you were trying to escape.
Data-driven computer models can solve problems you've wrestled with for years, but those models won't explain their reasoning. You're expected to trust the algorithm and hope for the best. And that's a tough sell when you're asking leadership to approve a significant investment in enzyme development.
What Physics-Based Modeling Actually Reveals in Enzymes
Here's an analogy that reflects the current state of enzyme design. Would you trust a pilot trained solely on the pattern recognition used by AI? Or would you rather fly with one who learned in a physics-based flight simulator?
The simulator models weight, speed, wind resistance, aerodynamics — the actual forces acting on the aircraft. When something goes wrong, the pilot understands why. They can adjust based on learned principles and not solely depend on what they encountered in previous flights.
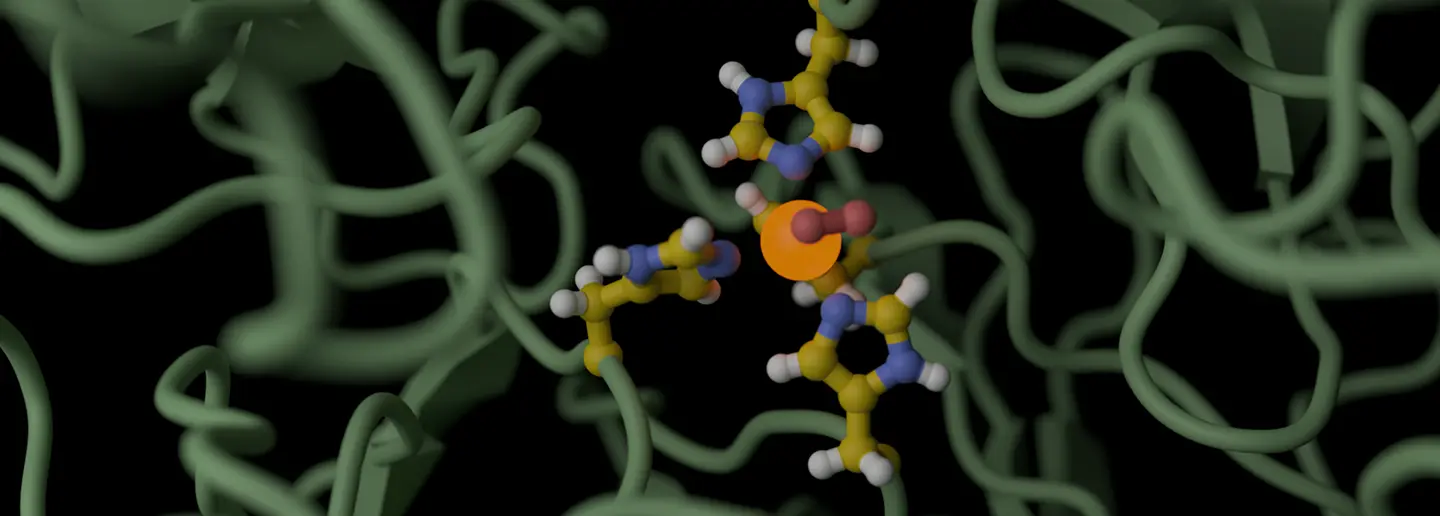
Physics-based enzyme modeling works the same way. Instead of learning from historical data, it simulates molecular reality. It calculates the enzyme-substrate encounters, conformational and allosteric effects, energetics, while tracking structural biology changes in real time.
Think about watching water molecules arrange themselves as ice forms. You can see the hydrogen bonds snap into place, witness the crystal structure emerge, and understand why ice floats based on observing molecular behavior. Physics-based modeling gives you that same visibility into enzyme catalysis — you're watching the reaction at the atomic level.
You can see binding modes. You can measure distances between key residues and track how a substrate accesses and fits into the active site, which interactions are favorable, which orientations drive the reaction forward. It's the difference between being told "this enzyme works" and seeing why it works. This interpretability is strategically essential to enzyme design.
Why Interpretability Protects Your Investment
When you're committing serious resources to enzyme development, interpretability gives you four critical advantages:
1. You can validate before you commit
A physics-AI hyperrealistic approach to modeling lets you screen millions of enzyme sequences and identify the candidates most likely to succeed. You're de-risking at the computational stage, reserving expensive lab work for variants with demonstrated viability.
Traditional directed evolution might test a million enzyme variants, but the vast majority are inactive or marginally improved clones that offer nothing useful. Companies that have spent decades accumulating this experimental data now face a paradox: They have massive datasets, but so much noise that extracting meaningful patterns becomes nearly impossible. Out of hundreds of thousands or even millions of historical data points, perhaps only 10 to 50 contain improved variants for training better models.
In contrast, a physics-AI approach to enzyme design might send fewer variants to the lab, but because they've been computationally screened for certain features, your hit rate is dramatically higher. In one project, Zymvol trained our AI and used just over 60 variants to outperform what 32,000 experimental clones had achieved. That's not luck. That's predictive accuracy driven by understanding.
2. You can troubleshoot with precision
When experimental results deviate from predictions, interpretable physics-AI hyperrealistic models let you diagnose the issue at the molecular level. Maybe the active site geometry isn't quite right. Maybe there's an unanticipated clash affecting substrate binding. Maybe pH conditions are altering protonation states in ways that reduce activity.
With this visibility, you can rationally design modifications. You're not starting over. You're iterating based on observed insight.
3. You can defend your position externally
Regulatory agencies and patent examiners demand clarity in your explanations. When you file for approval or IP protection, you need to demonstrate structure-activity relationships and justify stability claims. Interpretable models generate the data and visualizations required to support these arguments. AI-only predictions can't meet this standard.
4. You can learn and improve over time
Every experiment you run with an interpretable physics-AI hyperrealistic model feeds back into institutional knowledge. Rather than just collecting pass/fail data, you're building an understanding of how molecular features influence enzyme performance in your applications.
This creates a virtuous cycle. Your physics-based predictions generate high-quality experimental data, and that data trains more accurate AI models. Those models suggest better candidates for physics-based validation. The combination accelerates faster than either approach alone.
The Reality Check in AI-driven Enzyme Design
Here's something you need to know: your competitors using AI-first approaches are already running into walls.
Companies that launched in the past two years promising AI-driven enzyme solutions are quietly pivoting. They're talking more about proteins, less about enzymes. They're hedging. Because when you start testing AI predictions in the lab, you discover that enzymes are much harder than advertised.
It's easy to promise fast results. It's harder to deliver on complex chemistry when your model can't explain why its predictions work.
This doesn't mean AI is useless. It means AI alone isn't yet ready for enzyme-specific challenges. The data requirements are too steep and the complexity too high, and each enzyme-substrate complex is a unique problem to tackle.
How the Landscape Has Already Shifted in Enzyme Design
When we launched nine years ago, enzyme development was prohibitively expensive. You were looking at €1 million to €1.5 million and 12 to 18 months of work just to get a lab scale working enzyme. Only pharmaceutical companies with massive budgets could afford it.
That world is gone. Physics-based modeling transformed it.
Today, enzyme development serves food ingredient manufacturers, materials companies, textile producers—industries that would never have considered biocatalysis under the old cost structure. Enzyme development is no longer the massive struggle it used to be, because computational and experimental innovations have dramatically reduced timelines and cost.
So in a real sense, AI is arriving to disrupt a problem whose first bottleneck has already been solved years ago.
That doesn't mean there's no role for AI. In drug discovery, AI is already imagining novel small-molecule structures that physics-based methods wouldn't have explored, revealing chemical space humans would not intuitively design. This is also a current trend in biologics, where we’re starting to see novel protein folds that have no precedent in nature.
The same will happen with enzymes. Eventually, AI systems will be capable of generating entirely new enzyme architectures, not merely variants of natural scaffolds, and some of those designs will unlock breakthrough applications, including enzymes that tolerate extreme industrial stresses (temperature, pH, solvents) and enable chemical transformations that nature never evolved for.
But that's the future, and we’re working hard to set the foundation. Right now, if you need an enzyme that works in your process, under your conditions, a physics-AI approach to modeling is the technology that delivers.
What You're Really Choosing in a Physics-AI-based Approach
Enzymes are quickly becoming the modern way of doing chemistry. If you're not exploring enzyme solutions now, you'll be catching up while competitors pull ahead. But modern doesn't mean reckless. It means using the tools that actually work.
Physics-based modeling delivers interpretable, defensible results right now. AI enhances the approach as it matures. Together, they'll unlock possibilities we're only beginning to imagine.
The question is whether you're willing to invest in solutions that work now, or gamble on promises about tomorrow.
You might be interested in
Start your Evolution
Create new products and processes, adapt existing ones or develop completely new biochemistry. Zymvol is here to guide you in any stage of your journey.
Go to solutions